Automatic Extraction System for RNA and DNA
$58,000.00
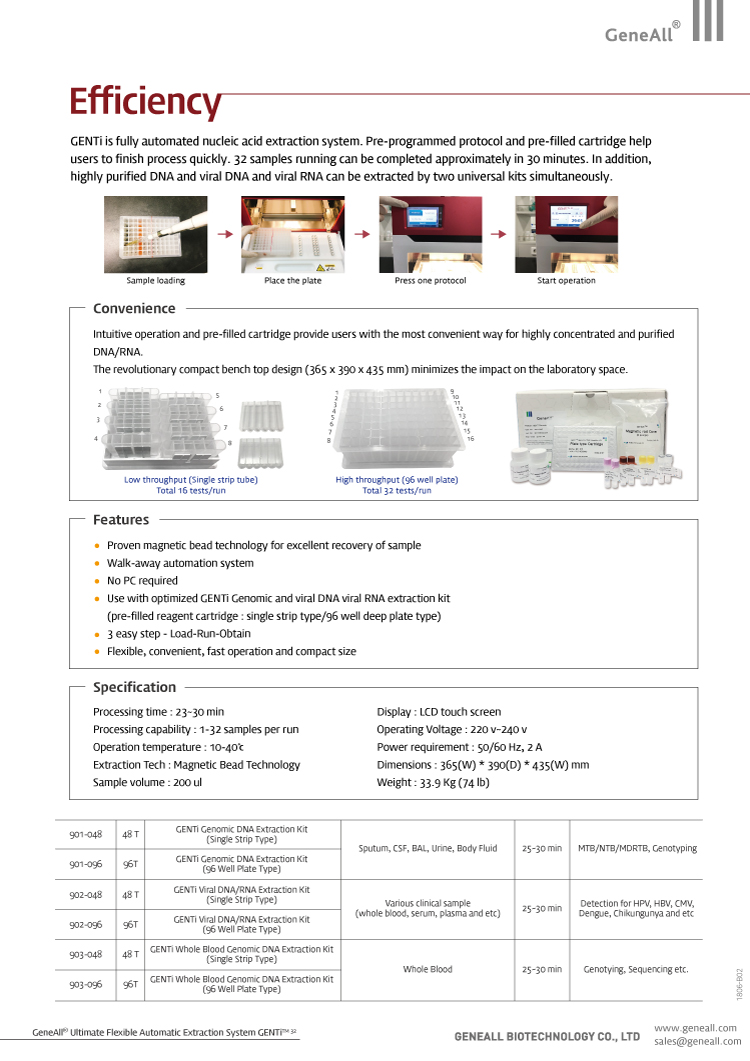
The GENTi™32 Automated Nucleic Acid Extraction system is a flexible and intuitive system that utilizes magnetic-bead technology to extract highly purified viral DNA and RNA from a a wide range of sample types.
| SKU | Stock | Units | Price | Quantity | ||
|---|---|---|---|---|---|---|
| GTI032 | Yes | 1 | $58,000.00 |
- Description
- Terms
- Additional information
- Product Details
- Documents
- Reviews (0)
Description
The GENTi™32 Automated Nucleic Acid Extraction system is a flexible and intuitive system that utilizes magnetic-bead technology to extract highly purified viral DNA and RNA from a a wide range of sample types. Extracted nucleic acid material is compatible with a wide range of downstream applications including PCR, qRT-PCR, RT-PCR, genotyping, gene expression, and sequencing. The automation allows for an instrument that requires ZERO specialized user training.
Only one manual step is required, the initial sample loading step, after which the entire process is fully automated eliminating potential errors in pipetting. To further ensure accurate results, the UV lamp can guarantee no risk of contamination.
Applications:
Automated Extraction of DNA and RNA for a variety of downstream applications including PCR, RT-PCR, qRT-PCR, sequencing, genotyping, and gene expression.
Key Features:
- Ultimate Flexibility: Under one operation protocol, all types of samples can be processed for nucleic acid extraction. Infinite capability for processing various types of samples: Two types of GENTi kits can process MOST types of samples
- Superior Processibility: 512 samples can be processed within 8 hours through 1 GENTi 32 (32 samples in 27 minutes)
- User-Friendly System: Single touch operation realized. All required chemicals, enzymes, and plastic consumables packed in a kit
- Two Configurations: SImulatneous operation with “single strip tube and 96-well plate”
Additional information
| Units | 1 |
|---|
Be the first to review “Automatic Extraction System for RNA and DNA”
You must be logged in to post a review.









Reviews
There are no reviews yet.